Image interpolation improves the zonal analysis of cartilage T2 relaxation in MRI
Introduction
Articular cartilage (AC), a thin layer of connective tissue coating the end of bones in joint, absorbs shock and distributes stress from the underlying bone. The major molecular components in cartilage are collagen, proteoglycan, and water. A unique morphological feature of AC that is relevant to this investigation is the depth-dependent properties of AC over its thickness, which is largely determined by its collagen orientation. Starting from its top surface (articular surface), cartilage is commonly subdivided into at least three structural zones, namely the superficial zone (SZ) in which the collagen fibers are aligned in parallel to the tissue surface, the transitional zone (TZ) where the fibers are in random orientation, and the radial zone (RZ) where fibers are perpendicular to the surface (1-3). Cartilage degradation causes the tissue to lose its load-bearing ability, which develops into the most common degenerative joint disease, osteoarthritis (OA) (4,5). Early detection of cartilage degeneration is the definitive yet unmet goal in clinical management of OA (6).
Most radiographic tools at the present time can only detect late stages of OA (7-10). Being non-invasive and sensitive to the molecular motions, magnetic resonance imaging (MRI) has undisputable potential to become the leading diagnostic tool for cartilage health (11-19). Several MRI protocols can in principle detect the early changes in OA cartilage, for example, T2 and T2 anisotropy, T1ρ, T1 with the use of contrast agent, and diffusion (20-25). The clinical method of OA detection using these MRI protocols, however, still needs further improvement. A major cause of this insufficient diagnosis is the relatively large voxel size in clinical MRI, in comparison with the thin thickness of AC over which the molecular concentration and orientation change significantly (6). Consequently, many properties that change naturally in the tissue (such as topographical distributions of the collagen orientation and proteoglycan concentration) can be masked by the changes of MRI parameters caused by the disease when the MRI resolution is insufficient.
In an ongoing multi-disciplinary imaging study of canine tibial cartilage that had surgically induced OA, we acquired T2 images of unopened canine knees at 200 µm pixel resolution. We then opened the knee joints and harvested the cartilage-bone specimens at the locations that were imaged previously, and imaged these specimens again at 17.6 µm pixel resolution. 2D T2 maps were calculated from both MRI experiments. In this project, we investigated if the reconstruction of the 200 µm images to the 100 µm images by image interpolation could improve the detection sensitivity by MRI T2 relaxation. Since the most important feature of AC is its depth-dependent zonal divisions, we aimed to investigate if different interpolating and zonal division methods could improve the detection of early OA.
Methods
Sample and procedure
This study used twelve mature canines, where six were non-operated controls and the other six each had the anterior cruciate ligament (ACL) transected on one of the knees 12 weeks prior to sacrifice. One knee from each of the six non-operated canines was labeled as healthy (N) (three left knees and three right knees). Six operated knees from six animals were labeled as OA and six non-operated adjacent knee were labeled as contralateral (C). These canine joints belonged to a multidisciplinary study of the OA progression in our lab, which were approved by the Canadian Council on Animal Care (CCAC). Recently, a blind study of the gross anatomic disease progression in these canine joints has been completed (26). Using the OARSI scores of 0–4 that graded different parts of a joint (27), the 12-week contralateral tibias had a score of 1, while the 12-week transected tibias (OA) had a score of 2; the difference was statistical significant. After excess tissues (muscles and skins) were trimmed, all unopened knee capsules were placed in a sample holder and imaged (within 24 hours of sacrifice) in a Varian MRI system with a 7T/20 cm horizontal magnet (Santa Clara, CA, USA)—the system is termed as macro-MRI in this report. After the imaging of the intact joints, each joint capsule was opened to harvest rectangular cartilage-bone blocks from the medial tibial plateau, with each block representing a different topographical location on the joint surface. Each specimen had an intact interface between cartilage and bone, and was approximately 3×3×4–5 mm3 in size. The blocks were immersed in the physiological saline solution, which also contained 1 mM in Gd-DTPA2– contrast agent (pertaining for the T1 study) and 1% protease inhibitor cocktail (Sigma, MO). The specimens were maintained at 4 °C (never frozen) and later imaged using µMRI.
MRI protocols
Quantitative T2 imaging in macro-MRI used a commercial multi-slice multi-echo (MSME) pulse sequence. The field of view (FOV) was set at 5 cm with a matrix size of 256×256, which yielded an approx. 200 µm pixel size. The repetition time was 3 sec with a total scan time of 51.2 minutes; and the ten echo times had an increment of 10 ms with the minimum echo time at 10 ms. Ten image slices, each having a thickness of 1 mm, were acquired, approx. 2.5 mm apart from each other. Microscopic MRI (µMRI) experiments on the specimens harvested from the same locations of the macro-MRI have been reported and were used as a reference (28-30). Briefly, quantitative T2 imaging used a magnetization-prepared imaging sequence on a Bruker AVANCE II system with a 7T/9 cm vertical magnet (Bruker Instrument, Billerica, MA, USA). The FOV was set as 0.45 cm and a slice thickness of 0.8 mm, resulting in a 2D pixel size of 17.6 µm. Other experimental details can be found in the papers cited above. T2 images for both macro-MRI and µMRI were calculated using a single exponential model using MATLAB (Mathworks, Natick, MA, USA) with a good correlation factor between data and model within the tissue region.
Interpolation of macro-MRI images
The native image in macro-MRI (256×256 matrix, with a nominal pixel size of 200 µm) was reconstructed to 512×512 (nominal pixel size of 100 µm) using two different methods. The first method (termed “FID 512”) was carried out on the Varian NMR spectrometer, utilizing the original time-domain Free Induction Decay (FID) data and the Fourier Transform reconstruction option (zero-filling). The second method (termed “ImageJ 512”) was carried out utilizing the image-domain 2D T2 images (256×256) and the bicubic interpolation procedure in a public domain software ImageJ® (version 1.4.3, NIH, MD, USA).
Division of sub-tissue in cartilage
The T2 profiles at both 200 µm/pixel and 100 µm/pixel were extracted from the 2D T2 images from five topographical locations on the medial tibia for each of the six healthy (N), contralateral and OA knees and also included the averaging of two adjacent columns to improve the signal to noise ratio (SNR). A count of sixty data points for each disease states resulted from six joints from five locations with two data pixels (ROI) per division. Since the column averaging was perpendicular to the direction of the cartilage depth, the depth resolution was still 200 or 100 µm. These T2 profiles were further analyzed using KaleidaGraph® (version 4.5.2, Synergy, PA, USA) by three methods of sub-tissue division. The first two methods divide the cartilage depth (thickness) into two equal-thickness divisions (upper 1/2 and lower 1/2), and three equal-thickness divisions (upper 1/3, middle 1/3 and lower 1/3), respectively. These equal-thickness division methods are similar to the common procedure found in clinical MRI of cartilage, since accurate morphological division of human cartilage is not easily available in clinical MRI due to the low pixel resolution. Our method divided the cartilage depth into three unequal-thickness zones (SZ, TZ, RZ), based on the average zonal thickness data from the known imaging and morphological findings at 17.6 µm resolution (28,30,31), with the first two pixels each representing the SZ and TZ respectively and the remaining pixels being assigned (as average) to the RZ. To resolve the issue of varying thicknesses across the selected tibial ROI, the T2 profiles were scaled at relative thickness with the articular surface (AS) as 0 and the bone-tissue interface as 1.
Statistical analysis
A one-way ANOVA with Tukey-Kramer HSD with α level of 0.017 (using JMP®) was performed on all specimens of the medial tibia, to pairwise compare T2 differences among healthy (N), contralateral (C) and OA samples at two different resolutions, with the three distinct types of division methods. For a highly effective result, a resultant P value of less than 0.01 was considered significant.
Results
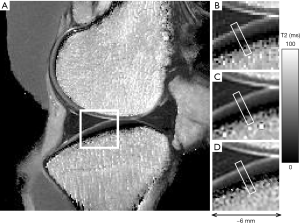
T2 map of a healthy medial tibia using the 7T macro-MRI imaged at the resolution of 200 µm/pixel (Figure 1A) showed the pixilation of a low-resolution whole joint image that is commonly seen in clinical MRI. The white region of interest (ROI) box depicted the region on the tibia from which a cartilage-bone specimen was later imaged at 17.6 µm/pixel in µMRI. Much like the original image (Figure 1B), similar ROIs were selected from the reconstructed images (100 µm/pixel) by two different types of interpolation methods: the ImageJ reconstruction (Figure 1C) and the FID reconstruction (Figure 1D). It is clear that both reconstruction methods produced smoother images (Figure 1C,D) that are lesser pixelated than the original image (Figure 1B) with minor differences in different regions of the joint. The smaller ROI within Figure 1B, C, and D depict ROI from which depth-dependent T2 profile analyses were performed and a similar location from which µMRI data were analyzed.
From the two-dimensional T2 maps, the depth-dependent T2 profiles for both healthy and OA cartilage (Figure 2A) were extracted from the ROI, with and without the image interpolation. Even though the images in Figure 1C and D had slight overall differences, the profiles showed near identicalness of the two interpolation methods as well the resolution difference between the original image and the interpolated images. The horizontal error bars in the original [256] T2 profile showed the pixel location at 200 µm and the site reference to the pixel locations at 100 µm. Since a 200 µm pixel is substantial in size when compared with the cartilage thickness (about 600–1,000 µm), the T2 value at 200 µm/pixel was placed in the middle of each large voxel in the horizontal scale (i.e., at the 100 µm location), as a representation of an average T2 value of the finite-sized voxel. For the 100 µm pixels, the T2 values were placed at the beginning, mid-point and end of each 256-pixel location. The most significant difference in T2 between healthy and OA cartilage is shown to occur near the top 20% of the tissue (Figure 2A). A careful inspection of the error bars and the comparison of the interpolated images using both methods showed that the time-domain (FID) reconstruction resulted in a slightly less noisy profile, and as such, was used for the rest of the analysis.
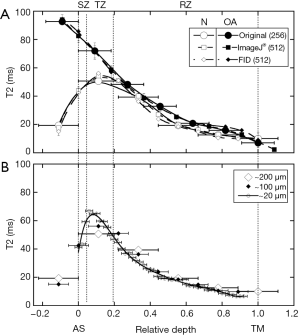
Comparison between the macro-MRI T2 profile of healthy cartilage (N), which were harvested from the ROI shown in Figure 1D, and their equivalent µMRI T2 profiles (Figure 2B) showed significant similarity (Figure 2B). The high-resolution µMRI data showed significantly more information in terms of the depth-dependency of cartilage T2 when compared to the macro-MRI data. The zonal markings on the µMRI profiles clearly indicated the resolution differences between the macro-MRI (~200 and 100 µm) and µMRI (~20 µm). This comparison demonstrated that the missing information in the low-resolution MRI is mainly in the top 20% of the cartilage in a typical clinical MRI setting. This missing information would depend on the ROI selection and the partial volume effect, which are critical for the early OA detection. The subtle yet critical difference between the 200 and 100 µm T2 profiles (N, healthy) where the 100 µm profile showed the lamina effect that is evident also in the 20 µm T2 profile (32,33). This difference is also documented in Table 1 in the unequal division of zone column (in italic) where the T2 pattern for 200 µm is SZ > TZ > RZ whereas the 100 µm follows a pattern of TZ > SZ > RZ, much similar to findings of healthy tissue T2 profiles in µMRI. This is a key finding in the importance of the reconstruction and minimum resolution needed for zonal division.
Full table
T2 profiles of the images at 200 µm/pixel resolution (Figure 3) and the “time-domain” interpolated images at 100 µm/pixel resolution (Figure 4) showed statistical findings among three different division methods (the two equal-thickness division, the three equal-thickness division, and the three unequal-thickness zones), and among three types of cartilage (healthy, contralateral, OA).

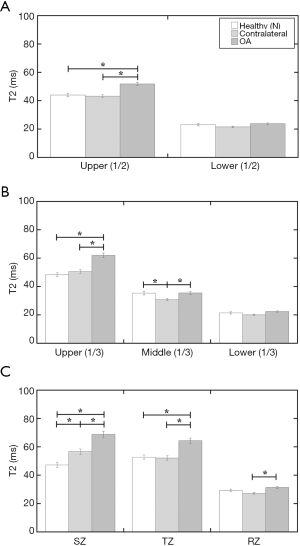
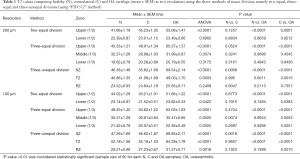
Table 1 summarizes the statistical findings (shown in Figures 3,4) as mean and standard error of the mean (SEM) with their respective statistical P values. Several conclusions can be drawn from the comparisons of the three division methods based on the strong statistical results.
- The subtle differences between the 200 µm/pixel data (Figure 3) and the 100 µm/pixel data (Figure 4) show that the image interpolation can improve the OA detection by MRI T2, even when the imaging resolution is still insufficient to resolve the structural zones in cartilage. This is best illustrated in the three equal-thickness division (Figure 4B), where the middle 1/3 gains T2 sensitivities to OA after image interpolation.
- Comparing the two and three equal-thickness division methods (Figures 3A/4A vs. Figures 3B/4B), it is clear that the three equal-division method at 100 µm resolution is more sensitive than the two equal-division—it is capable of picking up the differences between healthy and contralateral cartilage, as well as between contralateral and OA cartilage when the image interpolation is used. This shows the improvement of image resolution, in both acquisition and analysis, should be the top priority in clinical MRI development.
- The more revealing division method is the unequal-thickness zonal division, which requires the knowledge of the true zonal structure in cartilage, obtainable only by histology or µMRI. This unequal zonal division is capable of detecting the T2 changes not only between healthy and OA cartilage, but also between healthy to contralateral cartilage (Figures 3C and 4C), in both 200 and 100 µm/pixel images. The latter difference between healthy and contralateral cartilage in SZ cannot be seen in any of the equally divided regional analysis (Figures 3A,B and 4A,B) at both resolutions (200 and 100 µm). This is a significant finding.
Discussion
It is rare that one has the opportunity to study approximately the same cartilage specimens using both macro-MRI at 200 µm resolution and µMRI at 17.6 µm resolution, quantitatively, with the same MRI parameter (T2 relaxation), at the same field strength (7T), and with both healthy and well-characterized OA. Given our extensive experience with similar canine cartilage at microscopic resolutions (2,21,28,34), the multidisciplinary properties of this type of cartilage are well understood. Our study of canine tibial cartilage at 200 µm per pixel by macro-MRI has approximately the same length scale in comparison to the clinical MRI of a thicker human tibial cartilage at a fraction of millimeters per pixel using a whole-body MRI scanner, both resolving about three pixels across the thickness of cartilage based on the clinical settings implemented. Consequently, this animal MRI study and the common human MRI scans share a similar structural averaging in cartilage within each depth-dependent pixel, as well as the averaging between cartilage and its surrounding tissues (6). Since T2 is an important indicator in MRI study of cartilage damage due to OA (28,35,36), this project could help better illustrate the issues of MRI resolution at early stages of osteoarthritic cartilage.
Some of the early lesions in human OA can be localized near/around the surface of the tissue (37). The averaging of the surface tissue in diagnostic imaging could therefore hide vital information that is essential to the detection of early OA. Many clinical MRI analyses often compare just one bulk T2 value between the healthy and OA cartilage (3,38). With the improvement of clinical MRI technology, resolving cartilage thickness in 2–4 or more pixels becomes possible for quantitative human MRI (39). Consequently, these pixels of equal size could be used to represent different depths of cartilage (40,41). This type of depth-dependent regional analysis is superior to the bulk analysis, since a regional analysis separates the different characteristics between surface and deep cartilage. Since the combined thickness of both SZ and TZ is likely less than 1/3 of the total thickness, an equal thickness division essentially groups the complex surface structures of cartilage into one pixel, which could mask subtle changes of T2 in early OA (Figure 2). This project demonstrates clearly the additional detection sensitivity gained from the image interpolation, even when the original data is inadequate to resolve the zonal structure of cartilage. The approach in this animal cartilage study could be equally applied to clinical MRI of human cartilage, based on the scaling of imaging resolution in MRI (6).
This project has benefited from the knowledge from µMRI T2 profiles of the same cartilage (28-30), which clearly indicate the strong and distinct zonal profiles of cartilage T2 over its thickness, especially near the surface. Proper zonal division permits the best separation of superficial and transitional zones, which increases the sensitivity of the MRI protocol. This study therefore calls for the determination of the minimum resolution required for MRI of osteoarthritic cartilage. This determination, however, can be complicated in practice by the heterogeneity of the zonal structures in cartilage over a joint surface such as tibia or femur (42), as well as the dimension and orientation of the imaging voxel, which is commonly non-isotropic (2). By averaging only within each zone, a much better detection of the morphological changes occurring in early OA cartilage can be obtained.
In the post-acquisition image interpolation from the original 256 matrix to a 512 matrix, two different reconstruction methods showed minimal difference. This result is beneficial to most users since few could have repeated access to the original MRI system after the data acquisition. Since the scaling factor of two was used in both interpolation methods, the quantitative information from the interpolation is sufficiently within the limitations reported by other image interpolation studies where interpolation was shown a statistical improvement in structural changes and reduction of partial volume effect (43-45). For canine tibial cartilage, a minimum resolution of 100 µm/pixel is both necessary and sufficient, since µMRI results at a much higher resolution showed the thinnest zones were about 80–100 µm in thickness (46).
One methodological consideration in this data analysis is the location of the T2 values on the depth scale of cartilage. We placed the T2 values at the center of the pixels in the depth scale (at 100 µm, 300 µm, and so on). Arguments could be formulated to place the T2 value at the upper edge of the pixels (at 0 µm, 200 µm, and so on). Since the voxel size is large in comparison with the collagen structure in cartilage, the partial volume effect is an important factor when deciding the depth location of the first pixel of the tissue (47). We recommend that as long as the depth-dependent analysis approach is consistent, which merely represent a relative shift of the relaxation profile, the analysis should result in a similar conclusion. However, it does leave an ambiguity in the profile matching, as the selection is user dependent. Since the shape of a MRI voxel is commonly elongated (6), the averaging of the surface reflects the contribution not only from different cartilage but also some surrounding tissues. This is represented by the surface portion of the T2 profiles in Figure 2A, which include two possible scenarios, that is, whether the T2 values in the adjacent non-cartilage regions is lower or higher than the T2 values of the first cartilage pixel.
In conclusion, we demonstrate that interpolation of T2 images can better detect OA in cartilage, in the situation when the MRI resolution is insufficient to resolve the depth-dependent T2 characteristics in AC. Several zonal divisions of AC were compared using quantitative macro-MRI data. An unequal zonal division is shown to be much more statistically informative than the typical equal zone division and hence able to detect earlier signs of T2 changes due to OA. Since the relative length scale of canine cartilage and macro-MRI is nearly identical to those of human cartilage and whole-body MRI, this image analysis approach can benefit the clinical detection of early OA in humans.
Acknowledgements
The authors are grateful to Dr. James R. Ewing and Dr. Gary Ding at the Henry Ford Hospital Neurology Department, for their assistance to the Varian instrument.
Funding: Y Xia is grateful to the National Institutes of Health for the R01 grants (AR052353, AR069047).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The study was approved by institutional review board at University of Calgary on March 10, 2007, with an animal welfare assurance number of A5018-01 and written informed consent was obtained from all patients.
References
- Buckwalter J, Mow V. Cartilage repair in osteoarthritis. In: Moskowitz R, Howell D, Goldberg V, Mankin H. editors. Osteoarthritis: Diagnosis and Management. 2nd ed. Philadelphia: Saunders; 1992:71-107.
- Xia Y, Moody JB, Burton-Wurster N, Lust G. Quantitative in situ correlation between microscopic MRI and polarized light microscopy studies of articular cartilage. Osteoarthritis Cartilage 2001;9:393-406. [Crossref] [PubMed]
- Chang G, Xia D, Sherman O, Strauss E, Jazrawi L, Recht MP, Regatte RR. High resolution morphologic imaging and T2 mapping of cartilage at 7 Tesla: comparison of cartilage repair patients and healthy controls. MAGMA 2013;26:539-548. [Crossref] [PubMed]
- Spandonis Y, Heese FP, Hall LD. High resolution MRI relaxation measurements of water in the articular cartilage of the meniscectomized rat knee at 4.7 T. Magn Reson Imaging 2004;22:943-51. [Crossref] [PubMed]
- Turkiewicz A, Petersson I, Björk J, Hawker G, Dahlberg L, Lohmander L, Englund M. Current and future impact of osteoarthritis on health care: a population-based study with projections to year 2032. Osteoarthritis Cartilage 2014;22:1826-32. [Crossref] [PubMed]
- Xia Y. Resolution ‘Scaling Law’ in MRI of Articular Cartilage. Osteoarthritis Cartilage 2007;15:363-5. [Crossref] [PubMed]
- Vignon E, Piperno M, Le Graverand MP, Mazzuca SA, Brandt KD, Mathieu P, Favret H, Vignon M, Merle-Vincent F, Conrozier T. Measurement of radiographic joint space width in the tibiofemoral compartment of the osteoarthritic knee: comparison of standing anteroposterior and Lyon schuss views. Arthritis Rheum 2003;48:378-84. [Crossref] [PubMed]
- Buckland-Wright JC, Macfarlane DG, Williams SA, Ward RJ. Accuracy and precision of joint space width measurements in standard and macroradiographs of osteoarthritic knees. Ann Rheum Dis 1995;54:872-80. [Crossref] [PubMed]
- Ley CJ, Bjornsdottir S, Ekman S, Boyde A, Hansson K. Detection of early osteoarthritis in the centrodistal joints of Icelandic horses: Evaluation of radiography and low-field magnetic resonance imaging. Equine Vet J 2016;48:57-64. [Crossref] [PubMed]
- Hunter DJ, Altman RD, Cicuttini F, Crema MD, Duryea J, Eckstein F, Guermazi A, Kijowski R, Link TM, Martel-Pelletier J, Miller CG, Mosher TJ, Ochoa-Albiztegui RE, Pelletier JP, Peterfy C, Raynauld JP, Roemer FW, Totterman SM, Gold GE. OARSI Clinical Trials Recommendations: Knee imaging in clinical trials in osteoarthritis. Osteoarthritis Cartilage 2015;23:698-715. [Crossref] [PubMed]
- Conaghan P. Is MRI useful in osteoarthritis? Best Pract Res Clin Rheumatol 2006;20:57-68. [Crossref] [PubMed]
- Nemec U, Oberleitner G, Nemec SF, Gruber M, Weber M, Czerny C, Krestan CR. MRI versus radiography of acromioclavicular joint dislocation. AJR Am J Roentgenol 2011;197:968-73. [Crossref] [PubMed]
- Mosher TJ, Walker EA, Petscavage-Thomas J, Guermazi A. Osteoarthritis year 2013 in review: imaging. Osteoarthritis Cartilage 2013;21:1425-35. [Crossref] [PubMed]
- Matzat SJ, van Tiel J, Gold GE, Oei EH. Quantitative MRI techniques of cartilage composition. Quant Imaging Med Surg 2013;3:162-74. [PubMed]
- Wang Y, Teichtahl AJ, Cicuttini FM. Osteoarthritis year in review 2015: imaging. Osteoarthritis Cartilage 2016;24:49-57. [Crossref] [PubMed]
- Huang M, Schweitzer ME. The role of radiology in the evolution of the understanding of articular disease. Radiology 2014;273:S1-22. [Crossref] [PubMed]
- Choi JA, Gold GE. MR imaging of articular cartilage physiology. Magn Reson Imaging Clin N Am 2011;19:249-82. [Crossref] [PubMed]
- Pakin SK, Cavalcanti C, La Rocca R, Schweitzer ME, Regatte RR. Ultra-high-field MRI of knee joint at 7.0T: preliminary experience. Acad Radiol 2006;13:1135-42. [Crossref] [PubMed]
- Wang YX, Wang J, Deng M, Liu G, Qin L. In vivo three-dimensional magnetic resonance imaging of rat knee osteoarthritis model induced using meniscal transection. J Orthop Translat 2015;3:134-41. [Crossref]
- Smith HE, Mosher TJ, Dardzinski BJ, Collins BG, Collins CM, Yang QX, Schmithorst VJ, Smith MB. Spatial variation in cartilage T2 of the knee. J Magn Reson Imaging 2001;14:50-5. [Crossref] [PubMed]
- Alhadlaq HA, Xia Y, Moody JB, Matyas JR. Detecting structural changes in early experimental osteoarthritis of tibial cartilage by microscopic magnetic resonance imaging and polarised light microscopy. Ann Rheum Dis 2004;63:709-17. [Crossref] [PubMed]
- Newbould RD, Miller SR, Tielbeek JA, Toms LD, Rao AW, Gold GE, Strachan RK, Taylor PC, Matthews PM, Brown AP. Reproducibility of sodium MRI measures of articular cartilage of the knee in osteoarthritis. Osteoarthritis Cartilage 2012;20:29-35. [Crossref] [PubMed]
- Regatte RR, Akella SV, Lonner JH, Kneeland JB, Reddy R. T1rho relaxation mapping in human osteoarthritis (OA) cartilage: comparison of T1rho with T2. J Magn Reson Imaging 2006;23:547-53. [Crossref] [PubMed]
- Matzat SJ, Kogan F, Fong GW, Gold GE. Imaging strategies for assessing cartilage composition in osteoarthritis. Curr Rheumatol Rep 2014;16:462. [Crossref] [PubMed]
- Welsch GH, Mamisch TC, Hughes T, Zilkens C, Quirbach S, Scheffler K, Kraff O, Schweitzer ME, Szomolanyi P, Trattnig S. In vivo biochemical 7.0 Tesla magnetic resonance: preliminary results of dGEMRIC, zonal T2, and T2* mapping of articular cartilage. Invest Radiol 2008;43:619-26. [Crossref] [PubMed]
- Kahn D, Mittelstaedt D, Matyas J, Qu X, Lee JH, Badar F, Les C, Zhuang Z, Xia Y. Meniscus Induced Cartilaginous Damage and Non-linear Gross Anatomical Progression of Early-stage Osteoarthritis in a Canine Model. Open Orthop J 2016;10:690-705. [Crossref] [PubMed]
- Cook JL, Kuroki K, Visco D, Pelletier JP, Schulz L, Lafeber FP. The OARSI histopathology initiative - recommendations for histological assessments of osteoarthritis in the dog. Osteoarthritis Cartilage 2010;18 Suppl 3:S66-S79. [Crossref] [PubMed]
- Lee JH, Badar F, Matyas J, Qu X, Xia Y. Topographical variations in zonal properties of canine tibial articular cartilage due to early osteoarthritis: a study using 7-T magnetic resonance imaging at microscopic resolution. MAGMA 2016;29:681-90. [Crossref] [PubMed]
- Lee JH, Badar F, Kahn D, Matyas J, Qu X, Chen CT, Xia Y. Topographical variations of the strain-dependent zonal properties of tibial articular cartilage by microscopic MRI. Connect Tissue Res 2014;55:205-16. [Crossref] [PubMed]
- Lee JH, Badar F, Kahn D, Matyas J, Qu X, Xia Y. Loading-induced changes on topographical distributions of the zonal properties of osteoarthritic tibial cartilage--A study by magnetic resonance imaging at microscopic resolution. J Biomech 2015;48:3625-33. [Crossref] [PubMed]
- Lee JH, Xia Y. Quantitative zonal differentiation of articular cartilage by microscopic magnetic resonance imaging, polarized light microscopy, and Fourier-transform infrared imaging. Microsc Res Tech 2013;76:625-32. [Crossref] [PubMed]
- Xia Y, Farquhar T, Burton-Wurster N, Lust G. Origin of cartilage laminae in MRI. J Magn Reson Imaging 1997;7:887-94. [Crossref] [PubMed]
- Xia Y. Averaged and depth-dependent anisotropy of articular cartilage by microscopic imaging. Semin Arthritis Rheum 2008;37:317-27. [Crossref] [PubMed]
- Mittelstaedt D, Kahn D, Xia Y. Topographical and depth-dependent glycosaminoglycan concentration in canine medial tibial cartilage 3 weeks after anterior cruciate ligament transection surgery-a microscopic imaging study. Quant Imaging Med Surg 2016;6:648-60. [Crossref] [PubMed]
- Wyatt C, Guha A, Venkatachari A, Li X, Krug R, Kelley DE, Link T, Majumdar S. Improved differentiation between knees with cartilage lesions and controls using 7T relaxation time mapping. J Orthop Translat 2015;3:197-204. [Crossref]
- Wei B, Zong M, Yan C, Mao F, Guo Y, Yao Q, Xu Y, Wang L. Use of quantitative MRI for the detection of progressive cartilage degeneration in a mini-pig model of osteoarthritis caused by anterior cruciate ligament transection. J Magn Reson Imaging 2015;42:1032-8. [Crossref] [PubMed]
- Hwang WS, Li B, Jin LH, Ngo K, Schachar NS, Hughes GN. Collagen fibril structure of normal, aging, and osteoarthritic cartilage. J Pathol 1992;167:425-33. [Crossref] [PubMed]
- Ross KA, Williams RM, Schnabel LV, Mohammed HO, Potter HG, Bradica G, Castiglione E, Pownder SL, Satchell PW, Saska RA, Fortier LA. Comparison of Three Methods to Quantify Repair Cartilage Collagen Orientation. Cartilage 2013;4:111-20. [Crossref] [PubMed]
- Bangerter NK, Taylor MD, Tarbox GJ, Palmer AJ, Park DJ. Quantitative techniques for musculoskeletal MRI at 7 Tesla. Quant Imaging Med Surg 2016;6:715-30. [Crossref] [PubMed]
- Welsch GH, Mamisch TC, Marlovits S, Glaser C, Friedrich K, Hennig FF, Salomonowitz E, Trattnig S. Quantitative T2 mapping during follow-up after matrix-associated autologous chondrocyte transplantation (MACT): full-thickness and zonal evaluation to visualize the maturation of cartilage repair tissue. J Orthop Res 2009;27:957-63. [Crossref] [PubMed]
- Shao H, Chang EY, Pauli C, Zanganeh S, Bae W, Chung CB, Tang G, Du J. UTE bi-component analysis of T2* relaxation in articular cartilage. Osteoarthritis Cartilage 2016;24:364-73. [Crossref] [PubMed]
- Appleyard RC, Burkhardt D, Ghosh P, Read R, Cake M, Swain MV, Murrell GA. Topographical analysis of the structural, biochemical and dynamic biomechanical properties of cartilage in an ovine model of osteoarthritis. Osteoarthritis Cartilage 2003;11:65-77. [Crossref] [PubMed]
- Bernstein MA, Fain SB, Riederer SJ. Effect of windowing and zero-filled reconstruction of MRI data on spatial resolution and acquisition strategy. J Magn Reson Imaging 2001;14:270-80. [Crossref] [PubMed]
- Mayerhoefer ME, Szomolanyi P, Jirak D, Berg A, Materka A, Dirisamer A, Trattnig S. Effects of magnetic resonance image interpolation on the results of texture-based pattern classification: a phantom study. Invest Radiol 2009;44:405-11. [Crossref] [PubMed]
- Du YP, Parker DL, Davis WL, Cao G. Reduction of partial-volume artifacts with zero-filled interpolation in three-dimensional MR angiography. J Magn Reson Imaging 1994;4:733-41. [Crossref] [PubMed]
- Zhuang Z, Lee JH, Badar F, Xu J, Xia Y. The influences of different spatial resolutions on the characteristics of T2 relaxation times in articular cartilage: A coarse-graining study of the microscopic magnetic resonance imaging data. Microsc Res Tech 2016;79:754-65. [Crossref] [PubMed]
- Liu F, Chaudhary R, Block WF, Samsonov A, Kijowski R. Multicomponent T2 analysis of articular cartilage with synovial fluid partial volume correction. J Magn Reson Imaging 2016;43:1140-7. [Crossref] [PubMed]


